Researchers Aim to Analyse Pangenomes Using Quantum Computing
A new collaboration brings together a world-leading interdisciplinary team with skills across quantum computing, genomics and advanced algorithms. They aim to tackle one of the most challenging computational problems in genomic science: building, augmenting and analysing pangenomic datasets for large population samples. Their project sits at frontiers of research in both biomedical science and quantum computing.
The project, which involves researchers based at the University of Cambridge, the Wellcome Sanger Institute and EMBL’s European Bioinformatics Institute (EMBL-EBI), has been awarded up to US $3.5 million to explore the potential of quantum computing for improvements in human health.
The team aims to develop quantum computing algorithms with the potential to speed up the production and analysis of pangenomes – new representations of DNA sequences that capture population diversity. Their methods will be designed to run on emerging quantum computers. The project is one of 12 selected worldwide for the Wellcome Leap Quantum for Bio (Q4Bio) Supported Challenge Program.
Since the initial sequencing of the human genome over 20 years ago*, genomics has revolutionised science and medicine. Less than one per cent of the 6.4 billion letters of DNA code differs from one human to the next, but those genetic differences are what make each of us unique. Our genetic code can provide insights into our health, help to diagnose disease or guide medical treatments.
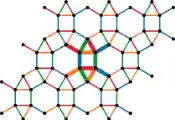
However, the reference human genome sequence, which most subsequently sequenced human DNA is compared to, is based on data from only a few people, and doesn’t represent human diversity. Scientists have been working to address this problem for over a decade, and in 2023 the first human pangenome reference was produced1. A pangenome is a collection of many different genome sequences that capture the genetic diversity in a population. Pangenomes could potentially be produced for all species, including pathogens such as SARS-CoV-2.
Pangenomics, a new domain of science, demands high levels of computational power. While the existing human reference genome structure is linear, pangenome data can be represented and analysed as a network, called a sequence graph, which stores the shared structure of genetic relationships between many genomes. Comparing subsequent individual genomes to the pangenome then involves mapping a route for their sequences through the graph.
In this new project, the team aims to develop quantum computing approaches with the potential to speed up both the key processes of mapping data to graph nodes, and finding good routes through the graph.
Quantum technologies are poised to revolutionise high-performance computing. Classical computing stores information as bits, which are binary – either 0 or 1. However, a quantum computer works with particles that can be in a superposition of different states simultaneously. Rather than bits, information in a quantum computer is represented by qubits (quantum bits), which could take on the value 0, or 1, or be in a superposition state between 0 and 1. It takes advantage of quantum mechanics to enable solutions to problems that are not practical to solve using classical computers.
However, current quantum computer hardware is inherently sensitive to noise and decoherence, so scaling it up presents an immense technological challenge. While there have been exciting proof of concept experiments and demonstrations, today’s quantum computers remain limited in size and computational power, which restricts their practical application. But significant quantum hardware advances are expected to emerge in the next three to five years.
The Wellcome Leap Q4Bio Challenge is based on the premise that the early days of any new computational method will advance and benefit most from the co-development of applications, software, and hardware – allowing optimisations with not-yet-generalisable, early systems.
Building on state of the art computational genomics methods, the team will develop, simulate and then implement new quantum algorithms, using real data. The algorithms and methods will be tested and refined in existing, powerful High Performance Compute (HPC) environments initially, which will be used as simulations of the expected quantum computing hardware. They will test algorithms first using small stretches of DNA sequence, working up to processing relatively small genome sequences like SARS-CoV-2, before moving to the much larger human genome.
Dr Sergii Strelchuk, Principal Investigator of the project from the Department of Applied Mathematics and Theoretical Physics, University of Cambridge2, said: “The structure of many challenging problems in computational genomics and pangenomics in particular make them suitable candidates for speedups promised by quantum computing. We are on a thrilling journey to develop and deploy quantum algorithms tailored to genomic data to gain new insights, which are unattainable using classical algorithms.”
David Holland, Principal Systems Administrator at the Wellcome Sanger Institute, who is working to create the High Performance Compute environment to simulate a quantum computer, said: “We’ve only just scratched the surface of both quantum computing and pangenomics. So to bring these two worlds together is incredibly exciting. We don’t know exactly what’s coming, but we see great opportunities for major new advances. We are doing things today that we hope will make tomorrow better.”
Dr David Yuan, Project Lead at EMBL-EBI, said: “On the one hand, we’re starting from scratch because we don’t even know yet how to represent a pangenome in a quantum computing environment. If you compare it to the first moon landings, this project is the equivalent of designing a rocket and training the astronauts. On the other hand, we’ve got solid foundations, building on decades of systematically annotated genomic data generated by researchers worldwide and made available by EMBL-EBI. The fact that we’re using this knowledge to develop the next generation of tools for the life sciences, is a testament to the importance of open data and collaborative science.”
The potential benefits of this work are huge. Comparing a specific human genome against the human pangenome – instead of the existing human reference genome – gives better insights into its unique composition. This will be important in driving forwards personalised medicine. Similar approaches for bacterial and viral genomes will underpin the tracking and management of pathogen outbreaks.